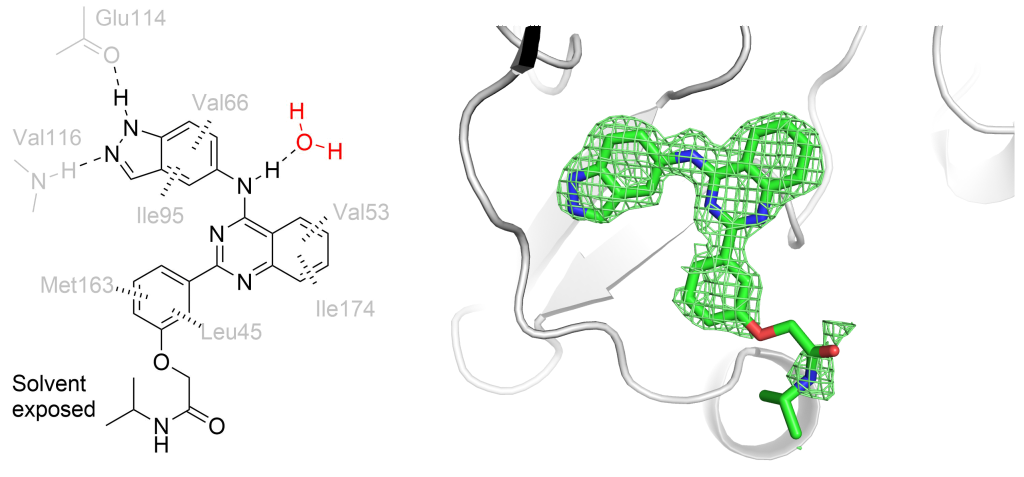
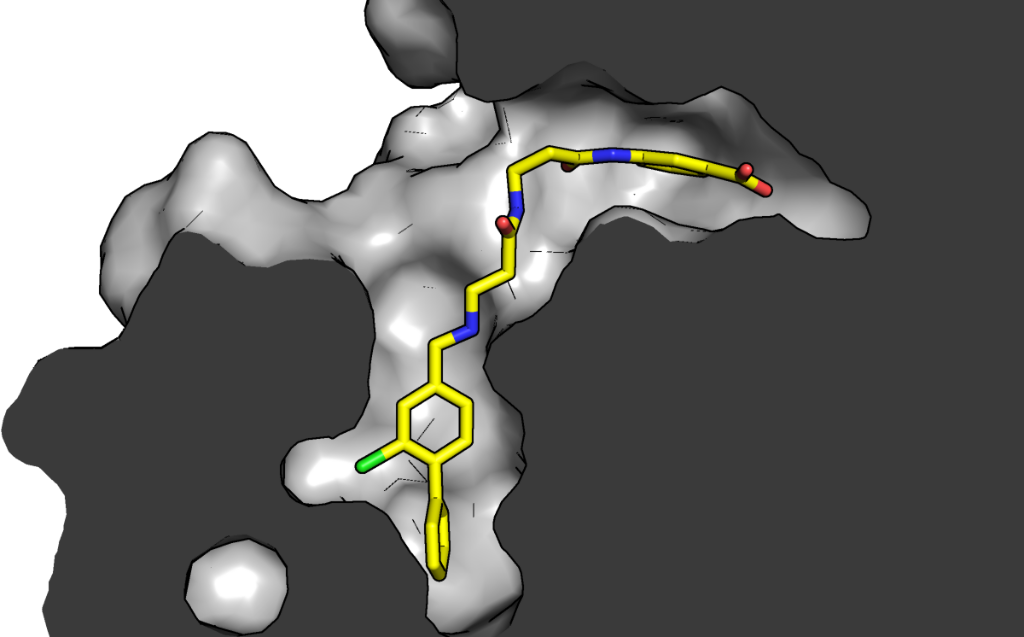
Dan Erlanson has highlighted our project on Ck2α inhibitor development in his Practical Fragments blog “Fragment Linking to selective Ck2 inhibitor“.
This a project we have done in close collaboration with David Spring’s group at the Department of Chemistry. The story started from a serendipitous observation that a fragment expected to bind on the top of the N-lobe of CK2α, on the interaction site with scaffolding protein Ck2β bound in a number of different sites on the kinase, including in a new pocket close to the ATP binding site of the kinase. This previously unidentified site was observed only thanks to a new crystal form Paul had obtained. In this crystal form, the so-called αD helix was mobile enough to be displaced by a fragment that, soaked at high concentration, found a new home in the very hydrophobic pocket that the displacement of the helix revealed. After some optimisation of the fragment and a crystallographic screen to identify ATP-site binding “war heads”, Claudia and others in the Spring lab were able to create the linked molecule CAM4066.
The main story was published in Chemical Science and the detailed description of the design process came out this year in Bioorganic and Medicinal Chemistry.
If interested more in this story, do check in Youtube some of the movies Paul has made from this project:
Development of CAM4066
Optimisation of the fragment in the aD pocket
Growth of the linker from the aD site to the active site
And it should not go forgotten that all the work has been guided continuous crystallographic assessement of the process, with ~30 unique crystal structures in the two papers: 5CVH (3D view), 5CVG (3D view), 5CVF (3D view), 5CU3 (3D view), 5CU4 (3D view), 5CU6 (3D view), 5CSH (3D view), 5CSP (3D view), 5CSV (3D view), 5CSH (3D view), 5CS6 (3D view), 5CLP (3D view), 5MMF (3D view ), 5MMR (3D view ), 5MO5 (3D view ), 5MO6 (3D view ), 5MO7 (3D view ), 5MO8 (3D view ), 5MOD (3D view ), 5MOE (3D view ), 5MOH (3D view ), 5MOT (3D view ), 5MOV (3D view ), 5MOW (3D view ), 5CU0 (3D view ), 5CU2 (3D view ), 5CT0 (3D view ), 5CTP (3D view ), 5CX9 (3D view ).
And there is more to come! Keep your eyes open.