Welcome to Sarah and Bocheng for joining the group for their part II projects (third year undergraduate project in normal terminology), under the supervision of Shiv and Emma, respectively.
Month: January 2023
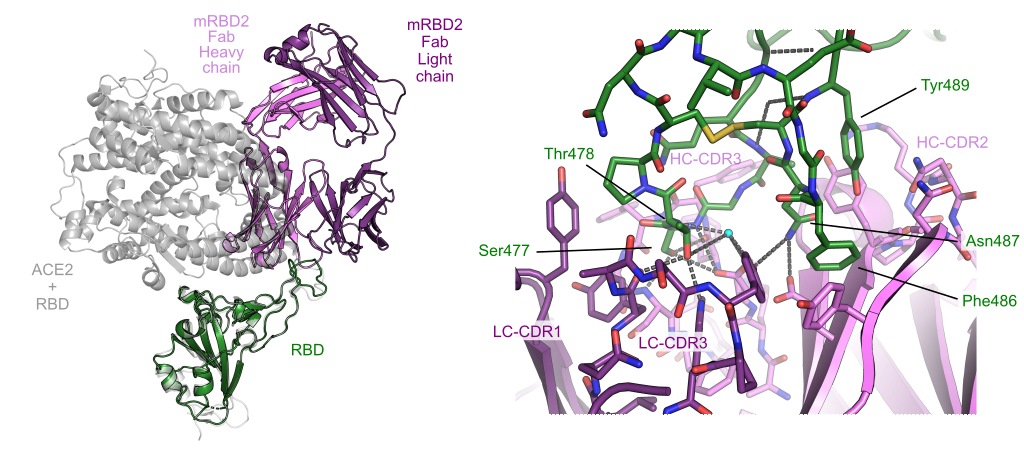
Microfluidics-enabled fluorescence-activated cell sorting of single pathogen-specific antibody secreting cells for the rapid discovery of monoclonal antibodies
Katrin Fischer, Aleksei Lulla, Tsz Y So, Pehuén Pereyra-Gerber, Matthew I. J. Raybould, Timo N. Kohler, Tomasz S. Kaminski, Juan Carlos Yam-Puc, Robert Hughes, Florian Leiß-Maier, Paul Brear, Nicholas J. Matheson,Charlotte M. Deane, Marko Hyvönen, James E. D. Thaventhiran, Florian Hollfelder
BioRxiv, posted 12 Jan 2023
DOI: 10.1101/2023.01.10.523494
PDB coordinates: 8BE1 (3D view)
Plasmids in Addgene: pExp-His-ZBasic-RBD, pExp-His-ZBasic-RBD-Avi
Abstract
Monoclonal antibodies are increasingly used to prevent and treat viral infections, playing a pivotal role in pandemic response efforts. Antibody secreting cells (ASCs, plasma cells and plasmablasts) are an excellent source of high-affinity antibodies with therapeutic potential. Current methodologies to study antigen-specific ASCs either have low throughput, require expensive and labour-intensive screening or are technically demanding and therefore not accessible to the wider research community. Here, we present a straightforward technology for the rapid discovery of monoclonal antibodies from ASCs: we combine microfluidic encapsulation of single cells into an antibody capture hydrogel with antigen bait sorting by conventional flow cytometry. Continue reading →
A versatile Halo- and SNAP-tagged BMP/TGFβ receptor library for quantification of cell surface ligand binding
Jerome Jatzlau, Wiktor Burdzinski, Michael Trumpp, Leon Obendorf, Kilian Roßmann, Katharina Ravn, Marko Hyvönen, Francesca Bottanelli, Johannes Broichhagen & Petra Knaus
Communications Biology 6: 34 (2023)
DOI: 10.1038/s42003-022-04388-4
Pubmed: 36635368
Abstract
TGFβs, BMPs and Activins regulate numerous developmental and homeostatic processes and signal through hetero-tetrameric receptor complexes composed of two types of serine/threonine kinase receptors. Each of the 33 different ligands possesses unique affinities towards specific receptor types. However, the lack of specific tools hampered simultaneous testing of ligand binding towards all BMP/TGFβ receptors. Here we present a N-terminally Halo- and SNAP-tagged TGFβ/BMP receptor library to visualize receptor complexes in dual color. In combination with fluorescently labeled ligands, we established a Ligand Surface Binding Assay (LSBA) for optical quantification of receptor-dependent ligand binding in a cellular context. We highlight that LSBA is generally applicable to test (i) binding of different ligands such as Activin A, TGFβ1 and BMP9, (ii) for mutant screens and (iii) evolutionary comparisons. This experimental set-up opens opportunities for visualizing ligand-receptor binding dynamics, essential to determine signaling specificity and is easily adaptable for other receptor signaling pathways.
