Background
The pBAT series of expression vectors have been constructed by Johan Peränen and co-workers at the Institute of Biotechnology, University of Helsinki and EMBL, Heidelberg
They differ from original pET vectors (developed initially by William Studier and co-workers) in some details (and these are the reasons why you should switch to BAT vectors ASAP!):
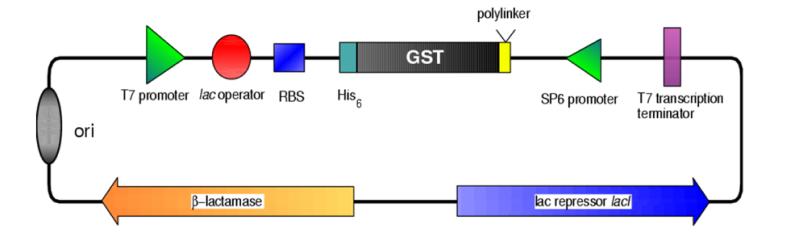
- The binding site for lac repressor is 1 nt closer to the T7 promoter than in pET vectors, hopefully allowing better repression.
- The origin of replication is derived from pUC vectors and thus the copy number of pBAT vectors is higher than pET vectors. Cloning is more pleasant while DNA yield from minipreps is very high. Higher copy number should also increase the number of repressor genes in the cell and subsequently the concentration of lacI repressor will be higher – again resulting in better repression of the T7lacpromoter
- The same difference also makes pBAT and its derivatives Bom– and hense non-mobilisable. pETs and other pBR322 derivatives (like pGEX) are Bom+ and hence only mobilisation defective. Gives you that extra bit of biosafety.
- SP6 promoter is located 3′ from the expressed gene allowing production of antisense RNA in case it is necessary. Also sequencing with SP6 primer is possible.
- The pBAT vectors are smaller than pET vectors.
- Various fusion vectors with compatible cloning sites are available, such as double affinity fusion vectors with His-tag followed by GST, MBP or TrxB fusion. This will facilitate purification and allows for example to overcome some annoying features of GST.
- The BAT vectors use NcoI as the standard restriction site for the 5′ end (with the initiation codon ATG). Unlike NdeI, which causes continuous misery for people by cutting poorly, ligating badly etc., NcoI is a pleasure to work with. And if NcoI would happen to cut your insert, there are many ways to overcome this problem.
- GST, MBP and thioredoxin fusions have been inserted intopHAT as SpeI fragments and can also be removed with the same enzyme to create a His-tag fusion with protease cleavage site (available separately as pHAT3 and pHAT4).
Note on vector maps: In most of the thrombin cleavable fusion vectors you will find two (2) BamHI sites: one immediately after the thrombin recognition sequence and one after the XhoI site. Although not unique, they are both shown in the vector maps in parentheses as the first one can be useful for 5′ cloning as long as NsiI or HindIII are used for the 3′ end.If you wish to check your mini/midi/maxi/whatever preps of original plasmids by restriction digest, I would recommend to use BspHI and HindIII.
When using these vectors, please cite publication Peränen et al., 1996, Analytical Biochemistry 236(2):371-3 (https://doi.org/10.1006/abio.1996.0187) .
The vectors
Basic vectors |
|||||
| pBAT4 | A non-fusion vector at the heart of the whole series. | ||||
| pBAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
| pBAT5 | A non-fusion vector without initiation ATG. | ||||
| pBAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
His-tag vectors |
|||||
| pHAT | Basic, non-cleavable N-terminal His-tag. | ||||
| pHAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
| pHAT2 | Minimal non-cleavable N-terminal His-tag. | ||||
| pHAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
| pHAT3 | Thrombin cleavable N-terminal His-tag. | ||||
| pHAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
| pHAT4 | Thrombin and TEV cleavable N-terminal His-tag. | ||||
| pHAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
| pHAT5 | C-terminal His-tag. | ||||
| pHAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
GST fusion vectors |
|||||
| pGAT | His-tag-GST fusion with a blunt end restriction site immediately after thrombin recognition sequence. |
||||
| pGAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
| pGAT2 | His-tag-GST fusion with a thrombin cleavage site. | ||||
| pGAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
| pGAT3 | His-tag-GST fusion with thrombin and TEV cleavage sites. | ||||
| pGAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
MBP fusion vectors |
|||||
| pMAT7 | His-tag-MBP fusion with blunt end restriction site immediately after thrombin sequence.. | ||||
| pMAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
| pMAT8 | His-tag-MBP fusion with a thrombin cleavage site. | ||||
| pMAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
| pMAT9 | His-tag-MBP fusion with a thrombin cleavage site and without NcoI site within MBP. | ||||
| pMAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
| pMAT10 | Like pMAT9, but without the second BamHI site in the polylinker | ||||
| pMAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
| pMAT11 | Like pMAT9, but with an additional TEV site | ||||
| pMAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
Thioredoxin fusion vectors |
|||||
| pTAT5 | His-tag-thioredoxin fusion with a thrombin cleavage site | ||||
| pTAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
| pTAT6 | His-tag-thioredoxin fusion with thrombin and TEV cleavage sites | ||||
| pTAT vector map (PDF) | Sequence: Staden, GCG , FASTA |
Restriction maps: All enzymes Unique sites |
|||
