Jessica Iegre, Paul Brear, Claudia De Fusco, Masao Yoshida, Sophie L. Mitchell, Maxim Rossmann, Laura Carro Santos, Hannah F. Sore, Marko Hyvӧnen and David R. Spring
Chemical Science (2018) (9:3041-)
DOI: 10.1039/C7SC05122K
Pubmed: 29732088
PDB coordinates:
6EHU (3D view ), 5OTQ (3D view ), 5OTY (3D view ), 6EHK (3D view ), 5OTI (3D view ), 5OTL (3D view ), 5OTO (3D view ), 5OYF (3D view ), 5OSZ (3D view ), 5OT5 (3D view ), 5OTD (3D view ), 5OTH (3D view ), 5OT6 (3D view ), 5OUE (3D view ), 5OUM (3D view ), 5OUU (3D view ), 5OS8 (3D view ), 5OTR (3D view ), 6EII (3D view ), 5OQU (3D view ), 5ORK (3D view ), 5OSL (3D view ), 5OUL (3D view ), 5ORH (3D view ), 5ORJ (3D view ), 5OS7 (3D view )

Abstract
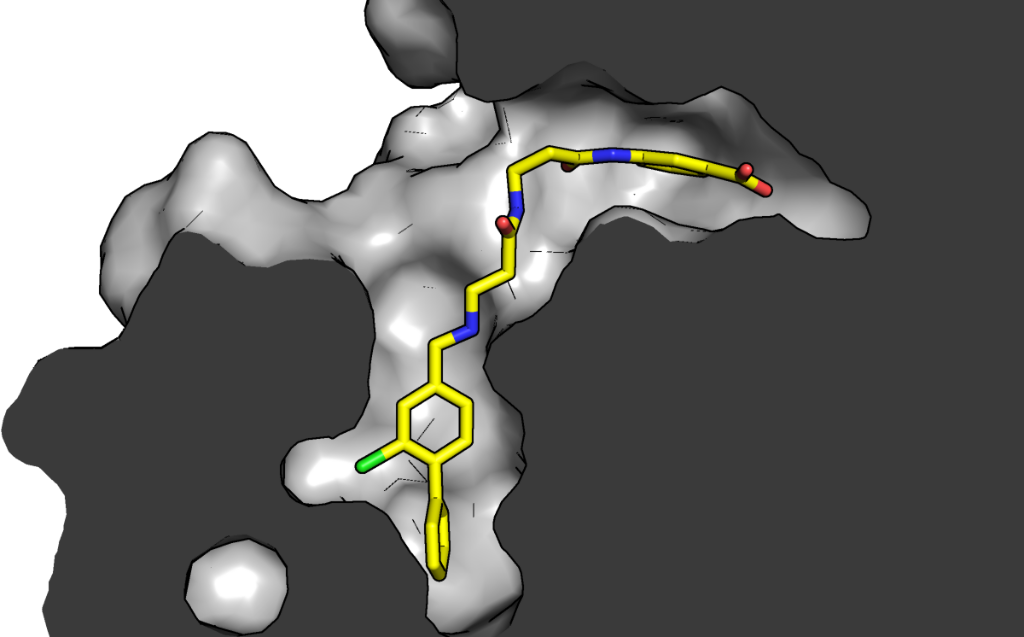
CK2 is a critical cell cycle regulator that also promotes various anti-apoptotic mechanisms. Development of ATP-non-competitive inhibitors of CK2 is a very attractive strategy considering that the ATP binding site is highly conserved among other kinases. We have previously utilised a pocket outside the active site to develop a novel CK2 inhibitor, CAM4066. Whilst CAM4066 bound to this new pocket it was also interacting with the ATP site: herein, we describe an example of a CK2α inhibitor that binds completely outside the active site. Continue reading →